Figure 2 reports that cis and trans regulatory divergences occur at similar frequencies, based on the activity of ∼10,000 regions per condition using ATAC-STARR-seq. However, the categorization into cis-only, trans-only, and cis-and-trans appears to heavily rely on arbitrary rank-based thresholds and assumptions of symmetric activity distribution across conditions.
This methodological decision raises a scientific concern:
To what extent do the predefined rank thresholds used to determine active regulatory regions bias the observed equivalence of cis and trans divergence frequencies?
Although the authors attempt to address robustness by comparing patterns across different thresholds (Figures S2L and S2M), it remains unclear whether the classification is driven by biological signal or artifacts of thresholding. Additionally, potential batch effects, differences in transfection efficiency, or subtle variations in chromatin accessibility between species may influence signal intensities and thus misclassify elements into divergence categories.
Is there any established methodology or published study that offers a more statistically robust framework to overcome this threshold-based bias in assigning cis and trans divergence?
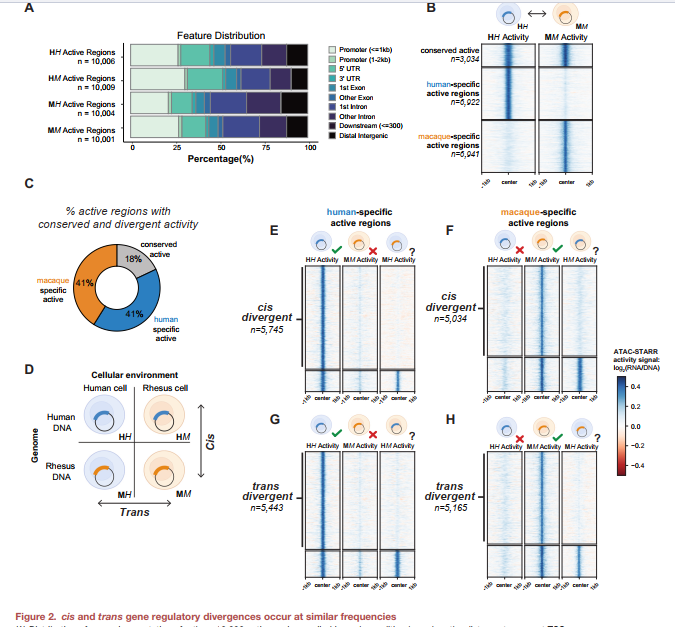


This concern is well founded, as threshold-dependent categorizations may introduce bias, especially in the context of high-throughput regulatory assays that involve inherent variability across replicates and conditions.
Several published methodologies offer more statistically rigorous alternatives to address this limitation. For example, Mattioli et al. (2020) employed a beta-binomial modeling framework to evaluate regulatory divergence, enabling probabilistic assessment of cis and trans effects while accounting for biological variability. This approach avoids rigid cutoffs and instead uses statistical confidence to define divergence categories more robustly.
10.1186/s13059-020-02110-3
Similarly, Naranjo et al. (2015) applied generalized linear models to dissect cis and trans regulatory contributions to gene expression, incorporating replicate-level data and enabling fine-scale partitioning of regulatory variation across species.
10.1371/journal.pgen.1005751
Bayesian hierarchical models or empirical Bayes approaches—such as those implemented in the limma or edgeR frameworks—may also offer more robust alternatives for analyzing regulatory activity by incorporating uncertainty and shrinkage estimators across experimental replicates.
In response to the central question: Yes, several established methodologies are available that overcome the limitations of hard thresholding by using statistically grounded frameworks. Applying such models to ATAC-STARR-seq or MPRA datasets can lead to a more accurate and biologically meaningful classification of cis and trans regulatory divergence.
Should there be interest in adapting these methods to a specific dataset or experimental design, further discussion or collaboration with computational genomics specialists could be valuable.